Whilst the planet soon learnt these few letters, mRNA, thanks to COVID and the lifesaving vaccine approach it afforded; RNA arguably is a molecular fossil – a piece of evolution that likely existed before modern cells.
The RNA world hypothesis suggests that life on Earth began with a simple RNA molecule that could copy itself without help from other molecules. How could we hypothesise this? Think about hereditary expression and how that is conferred. If this was indeed the case, then RNA would have to both store genetic information and catalyse the reactions for the cells. Curiously RNA persists in catalysing many of the fundamental processes in cells today. How DNA rose to be the genetic material and how did the code arise, when did proteins become the main catalyst? If we have such a little handle on the history, then the body of present knowledge is clearly lacking depth, a world of wonder is yet to be discovered. Insight can be found in “The RNA World and the Origins of Life”1
Understanding RNA
All living cells contain RNA (Ribonucleic Acid) in its many forms, whilst there are structural similarities to DNA there are clear differences. Most RNA is single stranded, has a backbone with alternating phosphate groups and a sugar called ribose. Each of these sugars has attached one of four bases, adenine (A), uracil (U), cytosine (C) or guanine (G). There are many forms of RNA such as: messenger RNA (mRNA), Transfer RNA (tRNA), ribosomal RNA (rRNA), Small Interfering RNA (siRNA), small RNA (sRNA) and the list seems to be expanding. RNA is ubiquitous. It is involved in gene expression and some viruses use it for their genomic material. RNA is a fundamental building block of life.
We are familiar with the use of mRNA for its usefulness in our immune responses to pathogens – such as COVID – 19. It’s success is largely due to a culmination of an enormous body of knowledge. Hundreds of scientists had worked on mRNA vaccines for decades before the coronavirus pandemic brought a breakthrough. Many Australian scientists including those at the Australian National University (ANU) made important contributions towards understanding the role of RNA. In the 1970s the Shine-Dalgarno sequence was discovered which tells the bacteria where to start protein synthesis so that the genetic code in mRNA is read correctly. This insight has enabled scientists to use bacteria as biofactories to make a host of different proteins that are now in use as drugs such as antibiotics, vaccines and cancer therapies or form part of important processes in biotechnology to develop yeasts, pesticides, enzymes, fuels, and solvents.
Traditionally, vaccines can take around 10 years to develop and consist of entire pathogens that have been killed or weakened so that they cannot cause disease. The recent COVID mRNA vaccines were developed in under a year and work by defining the genetic code of the target – easy now but before 1990 and the start of the Human Genome Project, this would have been particularly arduous. mRNA delivers the instructions your body needs to recognise the virus and fight it off. Cells then break down the mRNA and get rid of it. This gives cells the opportunity to change the type and number of proteins made based on demand which is key to allowing living things to grow, change, and react to their environment.
RNA is generally synthesised from DNA (often bacterial) by the enzyme RNA Polymerase through a method called transcription (think trans-scribe – i.e., to write) where not a copy but the complimentary RNA sequence to the DNA template is produced. The protein production is handled by the ribosome, these proteins are released, and the body sees them as foreign and mounts an antibody response, voila – a vaccine. No need to grow batches of cells in bioreactors or infect millions of eggs. The RNAs involved in this procedure are mRNA, tRNA and rRNA. In a self-amplified mRNA (samRNA) consider not only are we attempting to defeat a virus, but we also hijack another part of a virus’ genetic machinery to aid in it. The alpha virus is manipulated by replacing viral structural proteins with the gene of interest. The genes encoding the Alphavirus RNA replication machinery are retained which translates into an RNA Dependent RNA Polymerase that is responsible for creating many copies of the sub-genomic RNA, resulting in the translation of multiple antigens, thus reducing the initial dose requirements.
Plunge yourself back into the dark days of 2020, close your eyes and transport yourself to New York, picture the refrigerated trucks lined up with scores of dead as the morgues had overflowed, the palpable fear of everyone as they potentially could be carrying your death sentence. Then, as Sandra Lindsay (Director of nursing critical care at Long Island Jewish Medical Center) explained “My whole life just changed tremendously in that one moment in time,” she added “What was going through my mind is, I cannot wait for this needle to pierce my arm,” 2. Sandra was the first person in the U.S. to get a Covid vaccine outside a clinical trial. Interestingly Covid vaccines prevented more than 3.2 million deaths and 18.5 million hospitalisations in the U.S. from December 2020 through November 20222. In this context the mRNA vaccines were tremendous, they were developed and created in record time, it is likely the simplest, safest vaccine that has ever been produced. Such speed can only be appreciated in context, Moderna’s COVID vaccine development was initiated after the SARS-CoV-2 genome was posted on January 10, 2020; manufacture and delivery of clinical trials material was completed within 45 days, and the first trial participants were vaccinated on March 16, 2020, just 66 days after the genomic sequence of the virus was posted. Arguably a ‘vaccine’ material would have likely been produced in about a week – in contrast to a cell based or cultured vaccine would take months to produce, then the scale up to pandemic levels is another massive undertaking.
Bivalent vaccines that protect against 2 strains are commonplace now and have proven to show greater effectiveness. It was earlier postulated the possibility to load an array of vaccines into one shot – ponder having an annual shot for Covid, Flu and whatever else is lurking out there to mess with our day. RNA certainly lends itself to this, though vaccines are just one modality of use for RNA.
Things become less clear when defining the role of long non-coding RNAs (lncRNAs). Largely thought of as ‘Junk’ RNA in the past, the last decade has seen evidence mount for the lncRNAs3 having key roles in gene regulation and studies are noting a divergence of biogenesis of lncRNA compared to mRNA. Localisation of the lncRNA and their interactions with proteins, DNA and RNA appears to give insight into their roles be it interfering with signalling pathways, chromatin modulation, affecting stability and translation of mRNA in the cytoplasm along with the regulation and function of membraneless nuclear bodies. Such processes have a knock-on effect in gene expression impacting a varied array of physiopathological and biological conditions including cancer, immune responses, and neuronal disorders. Their localisation and condition specific patterns are gaining interest as biomarkers for disease states. So much for Junk!
RNA Therapeutics
Before we dive into what therapeutics are possible with RNA, perhaps it is best to understand how this can be achieved. Consider the process of DNA forming RNA then forming proteins – there are 3 distinct ways to use this process to prevent disease. I) Gene knockout – completely remove the DNA, 2) prevent or alter the Transcription of RNA from DNA, 3) prevent or alter the Translation of the protein.
Transcriptional Silencing is not as simple as once assumed. The most highly studied phenomenon in epigenetic modifications by far is DNA methylation, which typically refers to covalently attaching a methyl group (CH3) to the 5th position of the cytosine nucleotide by means of a group of specific enzymes called DNA methyltransferases (DNMTs) using S-adenosyl-L-methionine (SAM) as substrate4.
Resolving genetic defects at this point can have a “global” or complete rectification of the disorder, importantly other processes may be at play permitting alternative modes of action such as the terrific ‘R-loop’ research in Fragile X syndrome5.
One of the most important advances in biology has been the discovery that siRNA (small interfering RNA) is able to regulate the expression of genes, by a phenomenon known as RNAi (RNA interference). siRNA is a double stranded non-coding RNA typically 20 – 27 base pairs in length. It has some specific properties where the primary and secondary modes of action are the inhibition of translation and mRNA cleavage.
The major difference between siRNAs and microRNA/ miRNAs is that the former are highly specific with only one mRNA target, whereas the latter have multiple targets. microRNA controls gene expression mainly by binding with mRNA in the cell cytoplasm. Instead of being translated quickly into a protein, the marked mRNA will be destroyed and recycled, or it will be preserved and translated later. If miRNA is under expressed the level of the protein it normally regulates may be high, and visa versa6.
miRNA is often used in cancer diagnosis, cancer prognosis and drug discovery research given its use in determining the function of a protein or gene in a cell. The miRNA-based therapeutics could be categorised into two types: miRNAs mimics and miRNAs inhibitors. The former are double-stranded RNA molecules that mimic miRNAs, while the latter are single-stranded RNA oligos designed to interfere with miRNAs. For example, there are clinical trials into miRNA mimics to treat blood cancers, fibrosis, and a tumour suppressor miRNA for solid tumours just to show the breadth of application. For more about RNA therapeutics see Fig 1. from: RNA-based therapeutics: an overview and prospectus8
The term “undruggable” may be one day a distant relic in an era of genetic medicines. The growing understanding of RNA functions and their crucial roles in disease lends weight to broadening the therapeutic targets. Research is scratching the surface of what is possible and if the body of research into non-coding RNA is any indicator, this scratch will likely resolve any itch. Slack and Chinnaiyan’s review in Cell7 removes all doubt. RNA, due to its distinct physiological and physiochemical properties can theoretically target any gene of interest should the correct nucleotide sequence be selected. The enormity of the genome thus holds great prospect for such therapeutics, diagnostics, and silencers. Pitch this against the only 0.05% of the genome drugged by current approved protein targeted small molecule chemicals and antibodies. Besides, around 85% of proteins lack specific clefts and pockets for small molecules binding8.
The infancy of this technology and the runs on the board are encouraging. Fomivirsen is the first FDA-approved ASO ( antisense oligonucleotide) drug for treating cytomegalovirus retinitis (CMV) in patients with AIDS, Patisiran is the first FDA-approved RNAi-based drug for treating Familial amyloid polyneuropathy, also called transthyretin-related hereditary amyloidosis. Sarepta Therapeutics ‘Eteplirsen’ for Duchenne Muscular Dystrophy (DMD) has completed clinical trials. Notably the ASO for spinal muscular atrophy, Nusinersen targets the CNS (central nervous system) was approved in 2016, this previously undruggable disease has new hope. It is referred to as an orphan drug given the rare genetic nature of the disease. Showing promise was the Translate Bio MRT5005 inhalable mRNA treatment for Cystic Fibrosis. Unfortunately, there was no pattern of increases in ppFEV1 (percent predicted forced expiratory volume in 1 second) – a measure of lung function. Despite this, a great deal can be gleaned from this trial, notably the tolerability of multiple mRNA doses, the appeared absorption into the blood from an initial inhalation of the mRNA, plus evaluation of immunogenicity markers showed no clear pattern of anti-CFTR antibodies, anti-PEG antibodies or T-cell sensitisation to CFTR.
The mRNA was delivered in a LNP (Lipid NanoParticle) and promisingly there were no patients which had detectable levels of lipid in the blood10. Sanofi acquired Translate Bio in Aug 2021 post the announcement of the clinical trial results. Today, there are already 18 clinically approved RNA-based therapeutics, including two vaccines that made mRNA a household word during the COVID-19 pandemic11.
Initially RNA -based therapeutics were embraced with clear rationales for disease in oncology, neurology, and infections. Given the advancement in research there are around 8 ASOs, 3 siRNAs and two mRNA FDA approved drugs. Ito et al9 conducted a review of Clinical trials with ncRNAs showing a staggering 757,348 published articles with a PubMed search with 321,672 since January 2017. I mention this to articulate the depth of research. Of the 108 their exclusion criteria failed to remove, there is a clear trend, 95% were ncRNAs used as observational tools and clearly only a few interventional in clinical trial. I expect the number of interventional ncRNA therapeutics in Clinical trial to explode in the near future fuelled largely by small biotechs and academia spin outs.
RNA Diagnostics
The development of the Nobel Prize winning discovery of the Polymerase Chain Reaction (PCR) technique is an exemplar of how previous discoveries pave the path for something special. By 1980 all the components required to perform PCR amplification were known, it was only a matter of time before Kary Mullis put these pieces together to create the thermo-cycled PCR amplification we know today.
Reverse Transcription Polymerase Chain reaction (RT-PCR), Quantitative real-time Polymerase chain reaction (qPCR), Reverse Transcription quantitative real-time polymerase Chain reaction (RT-qPCR), and RNA sequencing (RNA-seq) today form the basis of RNA diagnostics. Whilst the application for RNA previously described here focusses on the production of a protein or a mechanism for gene manipulation, it is clear that RNA is dynamic with diverse and essential roles throughout the entire genome. The broad distribution and utility of the biomolecule has made it a foci for diagnostic, prognostic, and biomarker functions, however, the translation to clinical diagnostics has unearthed significant challenges, particularly in the realm of liquid biopsies.
Historically, the detection of specific mutations in cell free DNA (cfDNA) has been the main thrust of research with a few cfDNA tests being approved by the FDA for diagnosis and since 2020 only one is based on next generation sequencing. There currently are no cell free RNA (cfRNA) clinical diagnostic tests approved.
What is promising is the pot of gold that awaits the inventor of the technology to become the gold standard for diagnostics, the liquid biopsy market is expected to eclipse 5.8 billion dollars by 202612. Not withstanding the lack of standardisation of method to collect, prepare, screen and analyse a sample plus the contamination issues of cellular RNA and DNA the technology issue may be dwarfed by the enormity of the project. Firstly, consider the conundrum. There is a need to screen a population of otherwise asymptomatic individuals to gather the data required given the concept of cfRNA targeting is early detection – before a tumour is developed and there is cfDNA floating about. Historical retention samples are of little use currently as the modality of preservation has been shown to contaminate the results. Without standardised methods that respect the inherent biases and sufficient cohorts to assess, it is likely the requirements are beyond the grasp of even the most collaborative research projects. The data interpretation will be challenging and arduous for medical staff inadept in bioinformatics and statistical analysis. miRNA may be surpassed by long RNAs given some recent promising results in new disease-associated RNAs, albeit very early stages these may be the next revolution in screening and diagnosis.
CRISPR:
Better known as genetic scissors to the layperson, the discovery13 of this remarkable tool is a triumph for collaborative research, conference meetings, and a quest of deciphering the unknown. Bacteria have a mechanism for not only fighting off viruses but a memory method for future infections not only in the individual bacterium, but this memory is passed down through generations. This is an adaptive immune system that detects viral DNA and destroys it – to achieve this they use CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats). What is interesting is it is programmable, in the original study Doudna and Charpentier13 decided to use some purified Cas9 protein and combined them with a crRNA (Crispr RNA) strand to see if they could duplicate the phenomenon by finding a unique DNA sequence and cut it – this failed. Given the abundance of another RNA in bacteria and the vicinity of it to the CRISPR protein the collaborators postulated it may be needed. They included tracrRNA and this cleaved the DNA. Next step was to purify the tracrRNA and fuse it with the CRISPR, create an experiment where different genetic codes are programmed in to cut at specific lengths of DNA, this was hugely successful, earning a Nobel Prize.
An explosion of research to utilise this ensued, and the lay-media went for the unintended use of genetically engineering babies to be smarter, faster, stronger totally misinterpreting the intent of the research and it’s potential to be used to treat genetic diseases.
Putting the ethics aside, the use of RNA in this is pivotal. Using a guide RNA, CRISPR has transformed the world of genomics, now it is possible to target a disease such as Duchenne Muscular Dystrophy in the aim of resolving the genetic defect. CRISPR Cas9 can be used to disrupt a sequence- inactivating a gene, larger sections of DNA can be cleaved either side of the desired deletion followed by a cellular repair process that joins the strands thus deleting the gene.
Another action is to correct the gene by homology directed repair, the DNA is cleaved as in deletions but the cell uses the supplied DNA template to repair the break, thereby replacing the faulty DNA sequence or even inserting a new gene14.
‘In short, it’s only slightly hyperbolic to say that if scientists can dream of a genetic manipulation, CRISPR can now make it happen. At one point during the human gene-editing summit, Charpentier described its capabilities as “mind-blowing.” It’s the simple truth. For better or worse, we all now live in CRISPR’s world´15
Evolution of Nanomedicine Production Technology
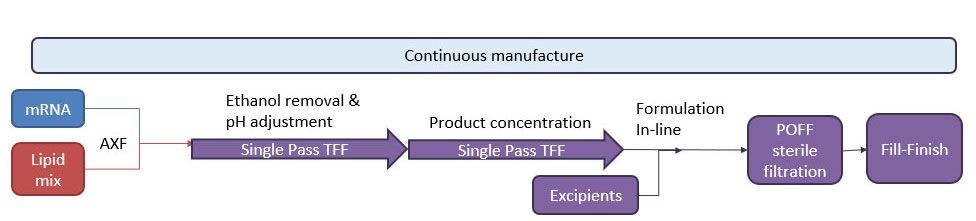
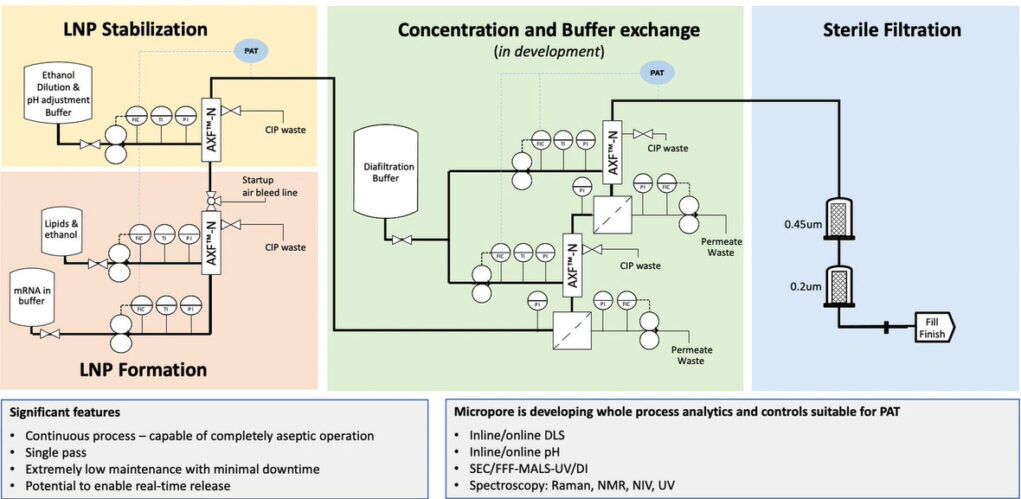
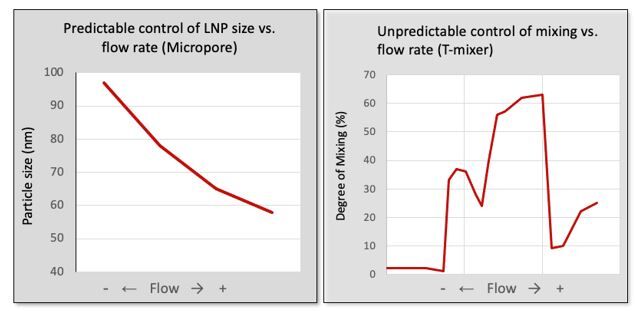
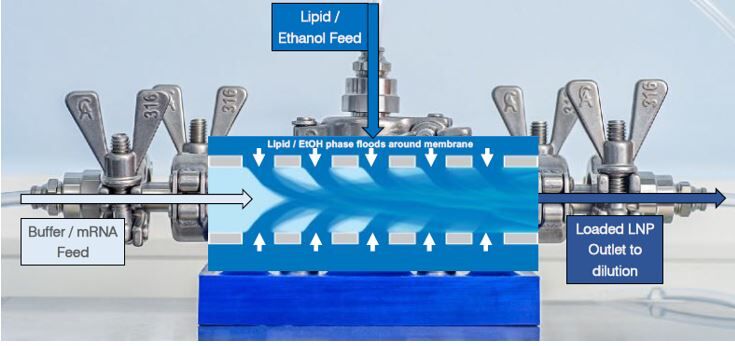
As RNA science continues to make headlines around the world as the new way of making safer, more targeted medicines, it has sparked a wave of new studies. With this comes the need for improved methods that deliver faster, more efficient and scalable drug manufacturing. Currently mRNA-LNP drugs are produced using methods such as lipid hydration, extrusion or impingement jet mixing (IJM), but these suffer from turbulent mixing and multiple harsh processing steps which compromise stability and give high batch variability. Microfluidic mixing offers rapid formulation with low polydispersity but again cannot accommodate high-volume production.
Micropore Pathfinder offers seamless scalability from initial R&D (0.2 mL) to final pandemic-scale GMP manufacturing (1500 L/h). This translates to 58,000 doses of vaccine every minute from a device small enough to fit in the palm of the hand. Collaborations with The University of Strathclyde and Professor Yvonne Perrie demonstrated mRNA encapsulation efficiencies over 97% in LNP production using Micropores AXF advanced crossflow mixing. This demonstrates that the Micropore Pathfinder can provide efficient mass production of a new generation of RNA-based therapeutics. The series is designed to be easy to operate, highly reproducible and stable in operation.
Book a demonstration with us today and try it yourself !
References.
- ‘The RNA World and the Origins of Life’ Bruce Alberts 2002; https://www.ncbi.nlm.nih.gov/books/NBK26876/ site access 15May2023
- ‘Two years after Covid vaccines rolled out, researchers are calling for newer, better options’ Aria Bendix, NBC News 14 Dec 2022. https://www.nbcnews.com/health/health-news/two-years-covid-vaccines-rcna57902 Site accessed 23 May 2023.
- ‘Gene regulation by long non-coding RNAs and its biological functions’ Luisa Statello et al 2020; https://www.nature.com/articles/s41580-020-00315-9 site accessed 17 May 2023
- ‘DNA Methylation Signature of Aging: Potential Impact on the Pathogenesis of Parkinson’s Disease’ Yazar V, Dawson VL, Dawson TM, Kang SU. 2023, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10041453/ site accessed 22 May 2023
- ‘Site-specific R-loops induce CGG repeat contraction and fragile X gene reactivation’ Hun-Goo Lee et al Cell, 2023. https://www.sciencedirect.com/science/article/abs/pii/S0092867423004695?dgcid=author Site accessed 23 May 2023.
- ‘The Limitless Future of RNA Therapeutics’ Damase et al ; 2021 https://www.frontiersin.org/articles/10.3389/fbioe.2021.628137/full Site accessed 24 May 2023
- ‘The Role of Non-coding RNAs in Oncology’ Frank J. Slack and Arul M. Chinnaiyan, Cell 2019. https://doi.org/10.1016/j.cell.2019.10.017 site accessed 25 May 2023
- ‘RNA-based therapeutics: an overview and prospectus’ Zhu et al; Cell death and Disease 2022. https://www.nature.com/articles/s41419-022-05075-2 Site accessed 25 May 2023
- “Current clinical trials with non-coding RNA-based therapeutics in malignant diseases: A systematic review’ Ito et al, Translational Oncology 2023. https://doi.org/10.1016/j.tranon.2023.101634 site accessed 25 May 2023
- Translate Bio Announces Results from Second Interim Data Analysis from Ongoing Phase 1/2 Clinical Trial of MRT5005 in Patients with Cystic Fibrosis (CF)’ Translate Bio, Inc. bit.ly/3BVUJC2 site accessed 25 May 2023
- ‘RNA therapeutics’ Michelle L. Hastings & Adrian R. Krainer; RNA (2023) Vol. 29, No. 4
- ‘Current challenges and best practices for cell-free long RNA biomarker discovery’ Cabús, L., Lagarde, J., Curado, J. et al. Biomark Res 10, 62 (2022) https://doi.org/10.1186/s40364-022-00409-w site accessed 30 May 2023.
- Discovery Story: Genome Engineering with CRISPR-Cas9 (Doudna, Jinek, Charpentier) https://youtu.be/jm5QqxN7Hkw iBiology.org May 2017 site visited 31 May 2023
- CRISPR/Cas9 – a specific, efficient and versatile gene-editing technology we can harness to modify, delete or correct precise regions of our DNA https://crisprtx.com/gene-editing/crispr-cas9 CRISPR Therapeutics 2023. Site accessed 31 May 2023.
And Science’s 2015 Breakthrough of the Year is… https://www.science.org/content/article/and-science-s-2015-breakthrough-year John Travis 2015. Site accessed 31 May 2023.




 02 9541 3500
02 9541 3500